|
|
|
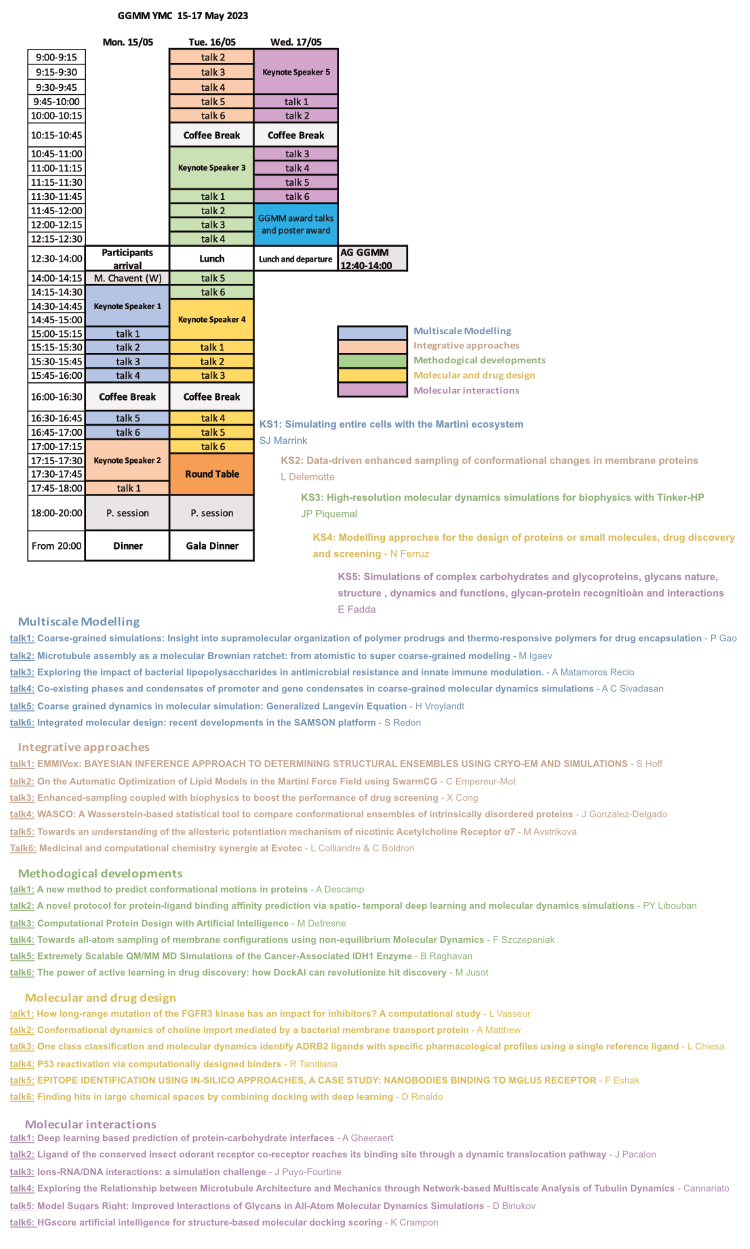
Program
The conference ebook is now available: here
This conference will be articulated around 5 topics (see below). For each topic, in addition of the keynote speaker, 5 talks (12 mns + 3 mns questions) will be selected from the abstracts. There will be also flash poster sessions so that each resercher will be able to briefly present (15-30s) his/her work to the audience before the poster session. Poster sessions will be held 15th and 16th evenings (from 6 to 8 pm). A round table (16th May, 5.15-6 pm) will be held to discuss career paths (in academia or private sector). This round table will involve keynote speakers and representatives of pharma/biotech companies.
Multiscale Moldelling: Modelling approaches combining different resolutions (QM, AT, CG, and beyond) applied on biological systems (protein, lipids nucleic acids, ..) and their interactions.
Pr SJ Marrink Simulating entire cells with the Martini ecosystem The ultimate microscope, directed at a cell, would reveal the dynamics of all the cell’s components with atomic resolution. In contrast to their real-world counterparts, computational microscopes are currently on the brink of meeting this challenge. In this talk, I show how, based on the Martini ecosystem, we are able to model an entire cell, the JCVI-syn3A minimal cell, at full complexity. This step opens the way to interrogate the cell’s spatio-temporal evolution with molecular dynamics simulations, an approach that can be extended to other cell types in the near future.
Integrative approaches: Approaches coupling modelling with data from bioinformatics (co-evolution, seq. alignments, …) or from experiments (structural biology, biophysics, …).
Pr L Delemotte Data-driven enhanced sampling of conformational changes in membrane proteins Membrane proteins engage in cellular communication and do so by cycling between various conformational states. Molecular dynamics simulations provide exquisite resolution insights into these processes but often are too short to be able to resolve full conformational cycles. In this talk, we will discuss how experimental data can be used to enhance the sampling of these conformational changes and construct free energy landscapes of membrane protein activation and gating. Comparing the landscapes obtained under different conditions (ligand-bound, pH, embedded in different membranes, etc...) help rationalize the effect of perturbations at the atomistic level.
Methodological Developments: Methodological developments to complete or go beyond MD simulations with a special interest to graphics, AI, and tools developments.
Pr JP Piquemal High-resolution molecular dynamics simulations for biophysics with Tinker-HP I will discuss our strategy for high-resolution molecular dynamics towards biophysical applications. As I detail the various protein targets that are currently under study, I will show how the newly developed multi-GPUs version of the Tinker-HP software [1,2] can accelerate high-resolution molecular dynamics simulations. Indeed, thanks to adaptive sampling and new generation many-body polarizable force fields such as AMOEBA, long (µs) molecular dynamics simulations at enhanced accuracy become possible.[3] As I detail the currently available other enhanced sampling capabilities of the software, I will give some perspectives about the use of new hybrid physically-driven machine learning approaches [4, 5] for condensed phase molecular dynamics. 1) Tinker-HP: a Massively Parallel Molecular Dynamics Package for Multiscale Simulations of Large Complex Systems with Advanced Polarizable Force Fields. L. Lagardère, L.-H. Jolly, F. Lipparini, F. Aviat, B. Stamm, Z. F. Jing, M. Harger, H. Torabifard, G. A. Cisneros, M. J. Schnieders, N. Gresh, Y. Maday, P. Ren, J. W. Ponder, J.-P. Piquemal, Chem. Sci., 2018, 9, 956-97 (Open Access), DOI: 10.1039/C7SC04531J 2) Tinker-HP: Accelerating Molecular Dynamics Simulations of Large Complex Systems with Advanced Point Dipole Polarizable Force Fields using GPUs and Multi-GPUs systems. O. Adjoua, L. Lagardère, L.-H. Jolly, Arnaud Durocher, Z. Wang, T. Very, I. Dupays, T. Jaffrelot Inizan, F. Célerse, P. Ren, J. Ponder, J-P. Piquemal, J. Chem. Theory. Comput., 2021, 17, 4, 2034–2053 (Open Access), DOI: 10.1021/acs.jctc.0c01164 3) High-Resolution Mining of SARS-CoV-2 Main Protease Conformational Space: Supercomputer-Driven Unsupervised Adaptive Sampling. T. Jaffrelot Inizan, F. Célerse, O. Adjoua, D. El Ahdab, L.-H. Jolly, C. Liu, P. Ren, M. Montes, N. Lagarde, L. Lagardère, P. Monmarché, J.-P.Piquemal, Chem. Sci., 2021, 12, 4889 – 4907 (Open Access), DOI: 10.1039/D1SC00145K 4) Scalable Hybrid Deep Neural Networks/Polarizable Potentials Biomolecular Simulations including long-range effects. T. Jaffrelot Inizan, T. Plé, O. Adjoua, P. Ren, H. Gökcan, O. Isayev, L. Lagardère, J.P. Piquemal, 2023, DOI: 10.48550/arXiv.2207.14276 5) Force-Field-Enhanced Neural Network Interactions: from Local Equivariant Embedding to Atom-in-Molecule properties and long-range effects. T. Plé, L. Lagardère, J.-P. Piquemal, 2023, DOI: 10.48550/arXiv.2301.08734 Molecular and Drug Design: Modelling approaches for the design of proteins (enzymes, ..) or small molecules, drug discovery and screening.
Dr N Ferruz De novo protein and enzyme design with unsupervised language models Artificial Intelligence (AI) methods are emerging as incredibly compelling tools in fields such as Natural Language Processing (NLP) and Computer Vision (CV), impacting the tools and applications we use in our daily lives. Language models have recently shown incredible performance at understanding and generating human text, producing text often indistinguishable from that written by humans. Inspired by these recent advances, we trained a language model, ProtGTP2, which effectively learned the protein language and generated sequences in unexplored regions of the protein space. A desirable critical feature in protein design is having control over the design process, i.e., designing proteins with specific properties. For this reason, we trained ZymCTRL, a model trained on enzyme sequences and their associated Enzymatic Commission (EC) numbers. ZymCTRL generates enzymes upon user-defined specific catalytic reactions, thus enabling conditional de novo design of biocatalysts. Our preliminary experimental data shows remarkable success, with high expression rates.
Molecular Interactions: Modelling biological complexes: protein-protein, protein-ligands, nucleic acids, sugars, …
Pr E Fadda Simulations of complex carbohydrates (glycans) and glycoproteins, glycans nature, structure, dynamics and functions, glycan-protein recognition and interactions Glucose is probably the most widely known sugar, generally recognized for its role in diet as the fundamental component of starch. Yet, the complexity of sugars in biology vastly exceeds cell metabolism, affecting virtually all aspects of the cell life cycle and of its interactions with the matrix, other cells and with pathogens, such as bacteria, toxins and viruses. The mystery surrounding sugars, less colloquially referred to as complex carbohydrates or glycans, derives from the fact that unlike proteins and nucleic acids, their biological synthesis is not template-driven and that they form highly flexible branched polymeric structures, ranging from one monosaccharide to systems counting hundreds of units. These two characteristics make them largely invisible in structural biology studies. Here I will discuss with you how high-performance computing can provide this missing fundamental information, allowing us to understand glycan structure and recognition, and thus their many biological functions in health and disease.
|